Introduction
We scanned the genomes of 8 organisms (A. thaliana, C. elegans, C. intestinalis, D. rerio, D. melanogaster, H. sapiens, M. musculus, and S. cerevisiae) with the JASPAR CORE PFMs associated with the same taxon to predict TFBSs and create the JASPAR TFBS genomic tracks. Moreover, we created a collection of familial TFBSs by merging overlapping TFBSs that were predicted from PFMs associated with the same familial binding profile. The TFBS predictions associated with all PFMs are available here.
We provide JASPAR TFBS predictions as genomic tracks, which can be visualized in genome browsers. Notably, the UCSC Genome Browser now presents predicted human JASPAR TFBS data as a native track for the human genome with information such as TF names, TFBS prediction scores, and PFM logo for each of the 12+ billion predictions.
The code and data used to create the UCSC tracks can be found at https://github.com/wassermanlab/JASPAR-UCSC-tracks.
Table of contents
UCSC tracks
Native tracks
Since the 2022 JASPAR release, the TFBSs are accessible as native UCSC genome tracks.
Familial binding sites tracks
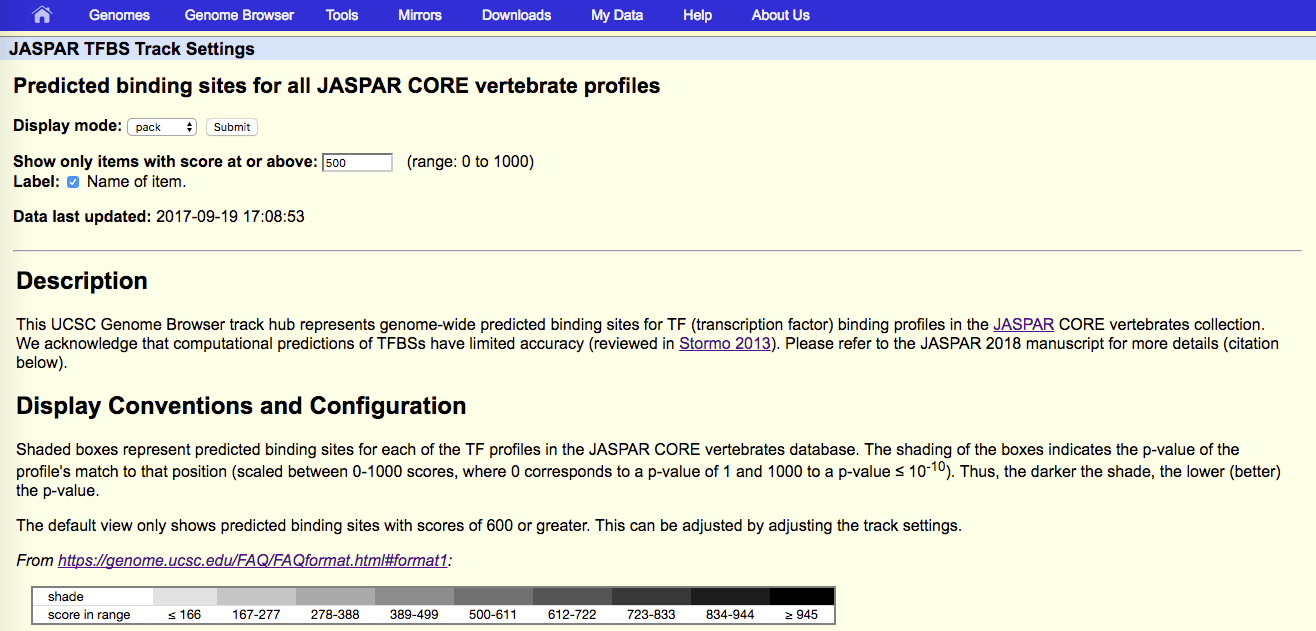
Genomic tracks can be viewed directly in the UCSC genome browser by connecting to the Public Hubs (Figure 1). The following genome assemblies are available and accessible directly by clicking on the assembly:
- Arabidopsis thaliana: araTha1
- Caenorhabditis elegans: ce10
- Caenorhabditis elegans: ce11
- Ciona intestinalis: ci3
- Danio rerio: danRer11
- Drosophila melanogaster: dm6
- Homo sapiens: hg19
- Homo sapiens: hg38
- Mus musculus: mm10
- Mus musculus: mm39
- Saccharomyces cerevisiae: sacCer3
Download
Underlying TFBS prediction data is available for download and further analysis: http://expdata.cmmt.ubc.ca/JASPAR/downloads/UCSC_tracks/
Example of dataset (random sample of lines):
chr1 280 298 AGL3 821 369 -
chr1 309 327 AGL3 823 373 +
chr1 309 327 AGL3 882 488 -
chr1 1577 1595 AGL3 823 373 +
chr1 1577 1595 AGL3 883 490 -
chr1 2113 2131 AGL3 853 428 +
chr1 3186 3204 AGL3 825 376 -
chr1 3543 3561 AGL3 834 392 +
chr1 4284 4302 AGL3 824 375 +
chr1 4302 4320 AGL3 838 399 -
chr1 4318 4336 AGL3 818 365 -
chr1 4386 4404 AGL3 826 378 -
chr1 4908 4926 AGL3 838 399 -
chr1 6799 6817 AGL3 811 354 -
chr1 7097 7115 AGL3 810 352 +
chr1 7339 7357 AGL3 829 383 -
chr1 8054 8072 AGL3 802 339 +
chr1 8054 8072 AGL3 815 359 -
chr1 8097 8115 AGL3 898 525 +
chr1 8097 8115 AGL3 945 665 -
Ensembl tracks
Direct link
View the tracks directly in the Ensembl Genome Browser using this link.
Attach track hub
You can attach the JASPAR track hub to Ensembl via “Configure this page”. Then search the Track Hub Repository for JASPAR and attach the hub.
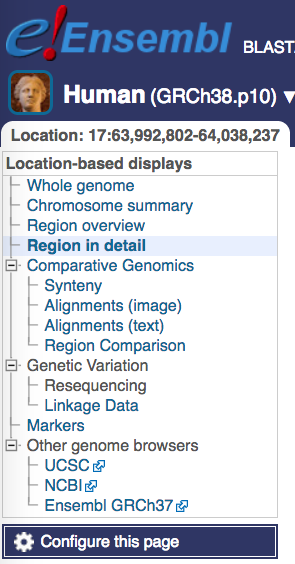
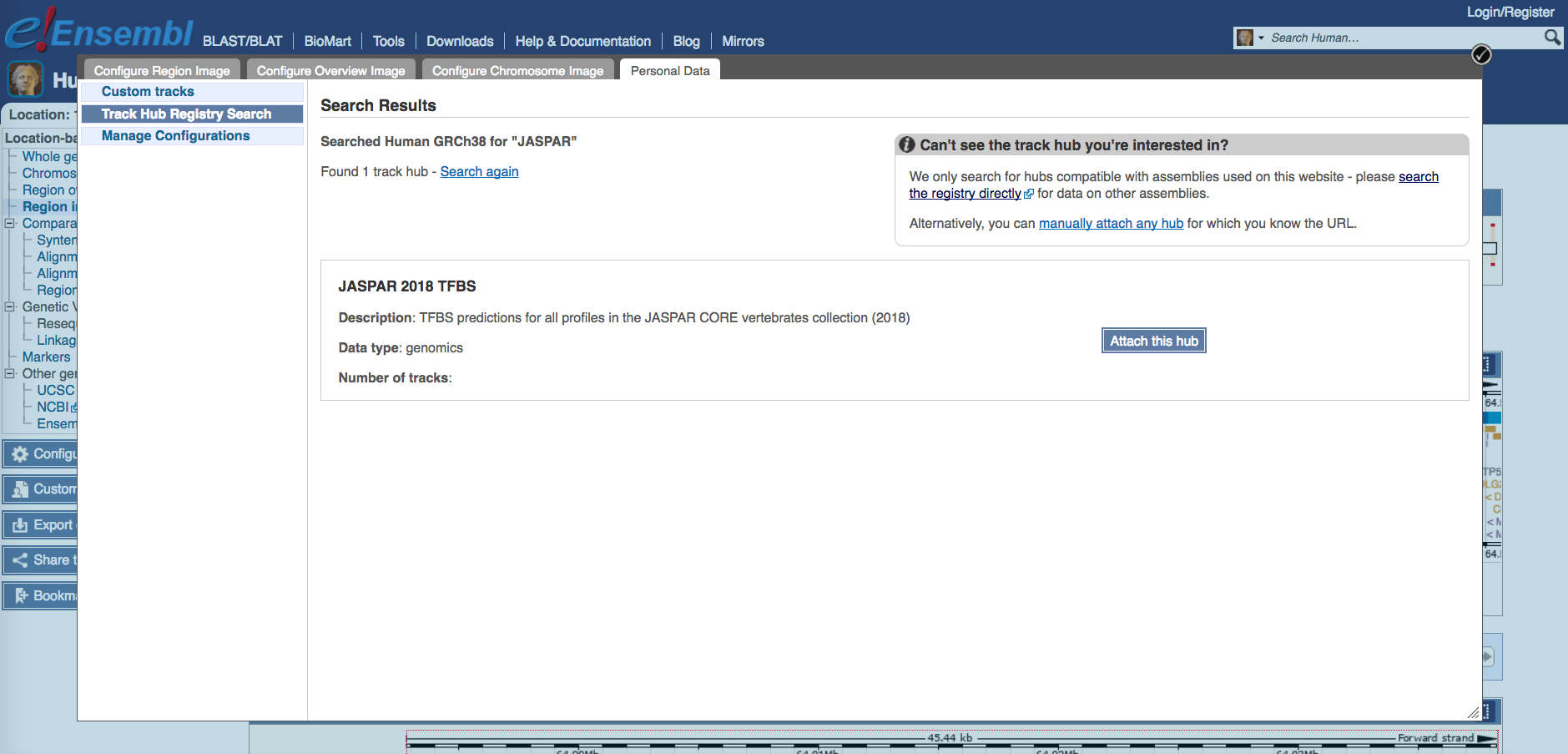
Note that while the JASPAR UCSC tracks by default show predicted TF binding sites with scores of 500 or greater (P < 10-5), the Ensembl tracks show all predicted binding sites. Ensembl colors predictions according to their score, from yellow (low scores) to green (medium) and blue (high). Please refer to the JASPAR hub help page (see below).
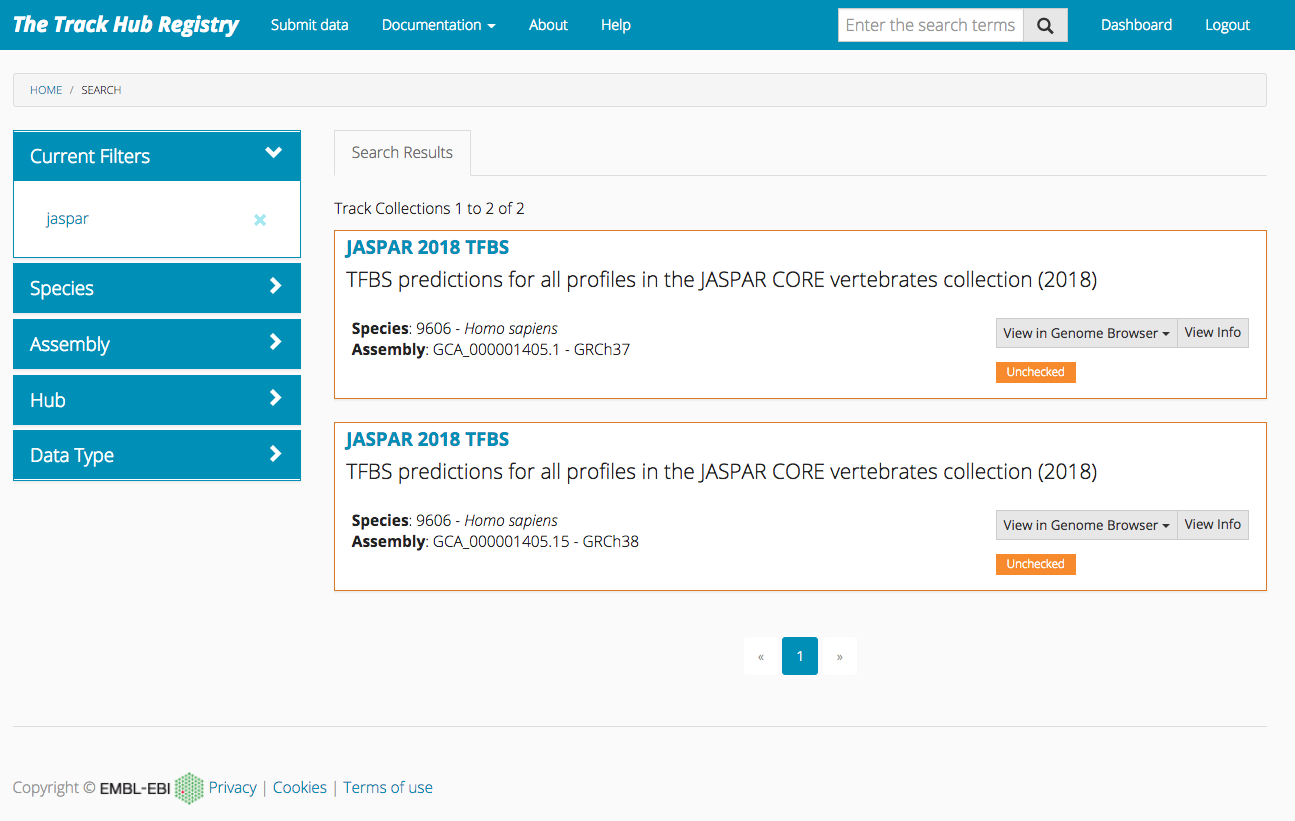
Track Hub Registry
JASPAR genome tracks are also available in Track Hub Registry. Use this link and select the track for the assembly of interest.
Further information
- Code and data used to create the UCSC tracks are available at https://github.com/wassermanlab/JASPAR-UCSC-tracks
- For more information on the use of UCSC track data hubs, see Using UCSC Genome Browser Track Hubs
- For more information on the use of track hubs in Ensembl, see http://www.ensembl.org/info/website/adding_trackhubs.html